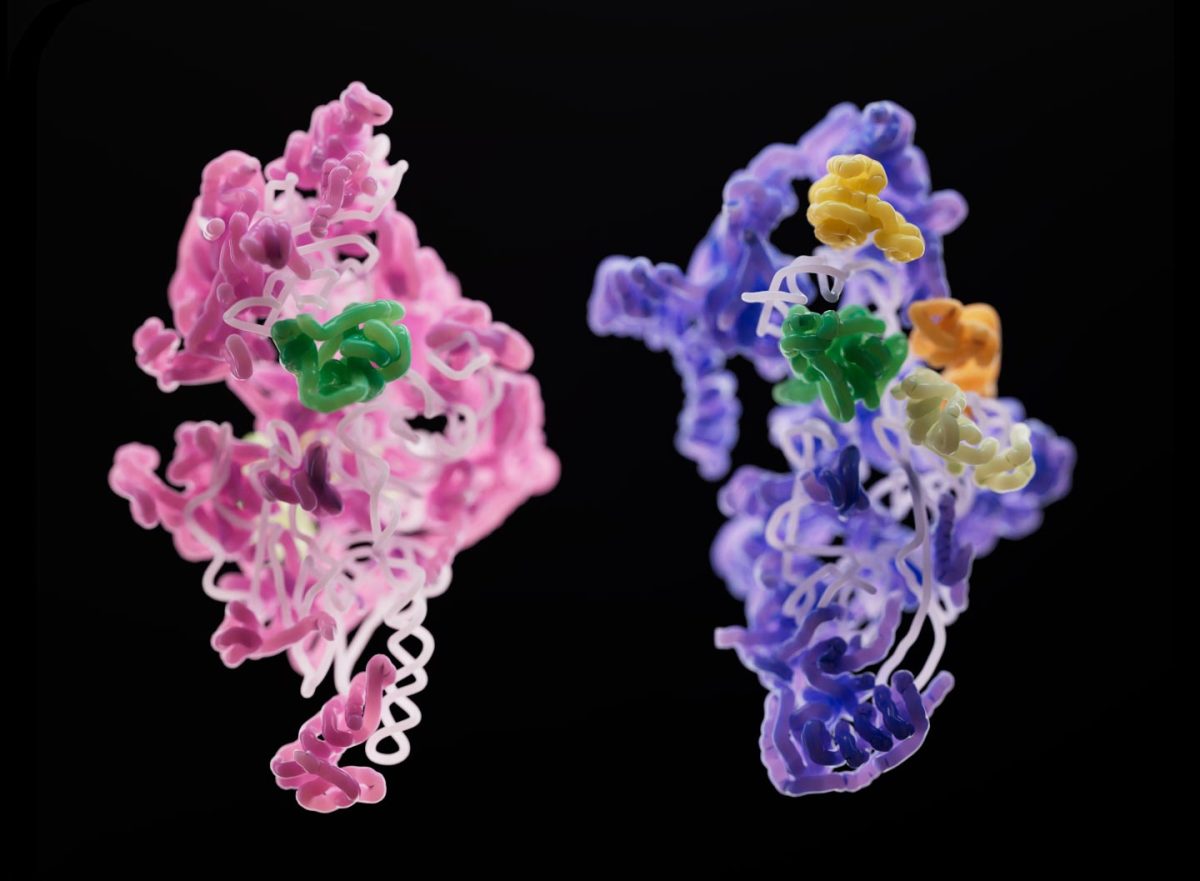
A subunit of a yeast mitoribosome (pink) compared to that of a human mitoribosome (purple). Although different, the two developing subunits have an assembly factor (green) in common. Credit: Sebastian Klinge
Across the tree of life, ribosomes, the tiny protein-producing factories within cells, are ubiquitous and look largely identical. Ribosomes that keep bacteria chugging along are, structurally, not much different from those churning out proteins in our own human cells.
But even two organisms with similar ribosomes may display significant structural differences in the RNA and protein components of their mitoribosomes. Specialized ribosomes within the mitochondria (the energy-producing entities within our cells), mitoribosomes help the mitochondria produce proteins that manufacture ATP, the energy currency of the cell.
The mitochondrial ribosome, or mitoribosome, is a protein complex that is active in mitochondria and functions as a riboprotein for translating mitochondrial messenger RNAs (mRNAs) encoded in mitochondrial DNA (mtDNA). The mitoribosome is attached to the inner mitochondrial membrane.
Scientists in the laboratory of Sebastian Klinge wondered how mitoribosomes evolved, how they assemble within the cell, and why their structures are so much less uniform across species. To answer these questions, they used cryo-electron microscopy to generate 3D snapshots of the small subunits of yeast and human mitoribosomes as they were being assembled. Their findings, which will be published today (December 8) in the journal Nature, shed light on the fundamentals of mitoribosome assembly, and may have implications for rare diseases linked to malfunctioning mitoribosomes.
“Three-dimensional pictures can tell us a lot about what steps are required, what proteins are involved in the process, and how you might be able to regulate the assembly of these large and complex machines,” says Nathan Harper, a graduate student in Klinge’s lab. “Cryo-EM allowed us to identify and isolate individual stages of the assembly pathway from a heterogeneous population of purified complexes, and we are able to see how these complexes change over time during assembly,” adds Chloe Burnside, also a graduate student in Klinge’s lab.
By observing this process in two different species—yeast and humans—the team managed to directly observe many similarities and differences in mitoribosome assembly. One key distinction: different proteins often were involved in otherwise similar acts of RNA folding. That’s likely because “there are common hurdles for these ribosomes,” Harper explains. “You can think about it like manufacturing two different bikes—a road bike and a mountain bike. You might need additional parts or tools for each one, but some key stages in production will be similar.”
The results provide unique insights into how molecular complexity and diversity arises in macromolecular complexes, and how assembly systems evolve along with the complexes themselves. A better understanding of mitoribosomes may also have implications for a range of severe diseases linked to mitoribosome dysfunction, such as Perrault syndrome. “We were able to map various disease-causing mutations onto different assembly factors’ structures, so that we could see how these mutations could affect the ribosome assembly process.”
Reference: “Principles of mitoribosomal small subunit assembly in eukaryotes” 8 December 2022, Nature.
DOI: 10.1038/s41586-022-05621-0
Share your story or advertise with us: Whatsapp: +2347068606071 Email: info@newspotng.com